Summary
Given a set of weighted features, identifies statistically significant hot spots, cold spots, and spatial outliers using the Anselin Local Moran's I statistic.
Learn more about how Cluster and Outlier Analysis (Anselin Local Moran's I) works
Illustration
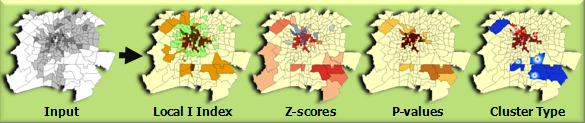
Usage
This tool creates a new Output Feature Class with the following attributes for each feature in the Input Feature Class: Local Moran's I index, z-score, pseudo p-value, and cluster/outlier type (COType).
The z-scores and p-values are measures of statistical significance which tell you whether or not to reject the null hypothesis, feature by feature. In effect, they indicate whether the apparent similarity (a spatial clustering of either high or low values) or dissimilarity (a spatial outlier) is more pronounced than one would expect in a random distribution. The p-values and z-scores in the Output Feature Class do not reflect any FDR (False Discovery Rate) corrections.
A high positive z-score for a feature indicates that the surrounding features have similar values (either high values or low values). The COType field in the Output Feature Class will be HH for a statistically significant cluster of high values and LL for a statistically significant cluster of low values.
A low negative z-score (for example, less than -3.96) for a feature indicates a statistically significant spatial data outlier. The COType field in the Output Feature Class will indicate if the feature has a high value and is surrounded by features with low values (HL) or if the feature has a low value and is surrounded by features with high values (LH).
The COType field will always indicate statistically significant clusters and outliers for a 95 percent confidence level. Only statistically significant features have values for the COType field. When you check the optional Apply False Discovery Rate (FDR) Correction parameter, statistical significance is based on a corrected 95 percent confidence level.
Default rendering for the Output Feature Class is based on the values in the COType field.
Permutations are used to determine how likely it would be to find the actual spatial distribution of the values you are analyzing. For each permutation, the neighborhood values around each feature are randomly rearranged and the Local Moran's I value calculated. The result is a reference distribution of values that is then compared to the actual observed Moran's I to determine the probability that the observed value could be found in the random distribution. The default is 499 permutations; however, the random sample distribution is improved with increasing permutations, which improves the precision of the pseudo p-value.
-
If the Number_of_Permutations parameter is set to 0, the result is a traditional p-value instead of a pseudo p-value and the z-score is based on the randomization null hypothesis computation. For more information on z-scores and p-values, see What is a z-score? What is a p-value?
When the Input Feature Class is not projected (that is, when coordinates are given in degrees, minutes, and seconds) or when the output coordinate system is set to a Geographic Coordinate System, distances are computed using chordal measurements. Chordal distance measurements are used because they can be computed quickly and provide very good estimates of true geodesic distances, at least for points within about thirty degrees of each other. Chordal distances are based on an oblate spheroid. Given any two points on the earth's surface, the chordal distance between them is the length of a line, passing through the three-dimensional earth, to connect those two points. Chordal distances are reported in meters.
When chordal distances are used in the analysis, the Distance Band or Threshold Distance parameter, if specified, should be given in meters.
-
For line and polygon features, feature centroids are used in distance computations. For multipoints, polylines, or polygons with multiple parts, the centroid is computed using the weighted mean center of all feature parts. The weighting for point features is 1, for line features is length, and for polygon features is area.
The Input Field should contain a variety of values. The math for this statistic requires some variation in the variable being analyzed; it cannot solve if all input values are 1, for example. If you want to use this tool to analyze the spatial pattern of incident data, consider aggregating your incident data. The Optimized Hot Spot Analysis tool may also be used to analyze the spatial pattern of incident data.
Your choice for the Conceptualization of Spatial Relationships parameter should reflect inherent relationships among the features you are analyzing. The more realistically you can model how features interact with each other in space, the more accurate your results will be. Recommendations are outlined in Selecting a Conceptualization of Spatial Relationships: Best Practices. Here are some additional tips:
- FIXED_DISTANCE_BAND
The default Distance Band or Threshold Distance will ensure each feature has at least one neighbor, and this is important. But often, this default will not be the most appropriate distance to use for your analysis. Additional strategies for selecting an appropriate scale (distance band) for your analysis are outlined in Selecting a fixed distance band value.
- INVERSE_DISTANCE or INVERSE_DISTANCE_SQUARED
When zero is entered for the Distance Band or Threshold Distance parameter, all features are considered neighbors of all other features; when this parameter is left blank, the default distance will be applied.
Weights for distances less than 1 become unstable when they are inverted. Consequently, the weighting for features separated by less than 1 unit of distance are given a weight of 1.
For the inverse distance options (INVERSE_DISTANCE, INVERSE_DISTANCE_SQUARED, or ZONE_OF_INDIFFERENCE), any two points that are coincident will be given a weight of one to avoid zero division. This assures features are not excluded from analysis.
- FIXED_DISTANCE_BAND
-
Additional options for the Conceptualization of Spatial Relationships parameter, including space-time relationships, are available using the Generate Spatial Weights Matrix or Generate Network Spatial Weights tools. To take advantage of these additional options, use one of these tools to construct the spatial weights matrix file prior to analysis; select GET_SPATIAL_WEIGHTS_FROM_FILE for the Conceptualization of Spatial Relationships parameter; and for the Weights Matrix File parameter, specify the path to the spatial weights file you created.
-
More information about space-time cluster analysis is provided in the Space-Time Analysis documentation.
-
Map layers can be used to define the Input Feature Class. When using a layer with a selection, only the selected features are included in the analysis.
If you provide a Weights Matrix File with a .swm extension, this tool is expecting a spatial weights matrix file created using either the Generate Spatial Weights Matrix or Generate Network Spatial Weights tools; otherwise, this tool is expecting an ASCII-formatted spatial weights matrix file. In some cases, behavior is different depending on which type of spatial weights matrix file you use:
- ASCII-formatted spatial weights matrix files:
- Weights are used as is. Missing feature-to-feature relationships are treated as zeros.
- If the weights are row standardized, results will likely be incorrect for analyses on selection sets. If you need to run your analysis on a selection set, convert the ASCII spatial weights file to an SWM file by reading the ASCII data into a table, then using the CONVERT_TABLE option with the Generate Spatial Weights Matrix tool.
- SWM-formatted spatial weights matrix file:
- If the weights are row standardized, they will be restandardized for selection sets; otherwise, weights are used as is.
- ASCII-formatted spatial weights matrix files:
Running your analysis with an ASCII-formatted spatial weights matrix file is memory intensive. For analyses on more than 5,000 features, consider converting your ASCII-formatted spatial weights matrix file into an SWM-formatted file. First put your ASCII weights into a formatted table (using Excel, for example). Next, run the Generate Spatial Weights Matrix tool using CONVERT_TABLE for the Conceptualization of Spatial Relationships parameter. The output will be an SWM-formatted spatial weights matrix file.
The Output Feature Class is automatically added to the table of contents with default rendering applied to the COType field. The rendering applied is defined by a layer file in <ArcGIS>/Desktop10.x/ArcToolbox/Templates/Layers. You can reapply the default rendering, if needed, by importing the template layer symbology.
-
The Output Feature Class includes a SOURCE_ID field which allows you to Join it to the Input Feature Class, if needed.
-
The Modeling Spatial Relationships help topic provides additional information about this tool's parameters.
When using this tool in Python scripts, the result object returned from tool execution has the following outputs:
Position Description Data Type 0
Output Feature Class
Feature Class
1
Index field name
Field
2
ZScore field name
Field
3
Probability field name
Field
4
COType field name
Field
5
Source ID field name
Field
Syntax
ClustersOutliers_stats (Input_Feature_Class, Input_Field, Output_Feature_Class, Conceptualization_of_Spatial_Relationships, Distance_Method, Standardization, {Distance_Band_or_Threshold_Distance}, {Weights_Matrix_File}, {Apply_False_Discovery_Rate__FDR__Correction}, {Number_of_Permutations})| Parameter | Explanation | Data Type |
Input_Feature_Class | The feature class for which cluster and outlier analysis will be performed. | Feature Layer |
Input_Field | The numeric field to be evaluated. | Field |
Output_Feature_Class | The output feature class to receive the results fields. | Feature Class |
Conceptualization_of_Spatial_Relationships | Specifies how spatial relationships among features are defined.
| String |
Distance_Method | Specifies how distances are calculated from each feature to neighboring features.
| String |
Standardization | Row standardization is recommended whenever the distribution of your features is potentially biased due to sampling design or an imposed aggregation scheme.
| String |
Distance_Band_or_Threshold_Distance (Optional) | Specifies a cutoff distance for Inverse Distance and Fixed Distance options. Features outside the specified cutoff for a target feature are ignored in analyses for that feature. However, for Zone of Indifference, the influence of features outside the given distance is reduced with distance, while those inside the distance threshold are equally considered. The distance value entered should match that of the output coordinate system. For the Inverse Distance conceptualizations of spatial relationships, a value of 0 indicates that no threshold distance is applied; when this parameter is left blank, a default threshold value is computed and applied. This default value is the Euclidean distance that ensures every feature has at least one neighbor. This parameter has no effect when Polygon Contiguity or Get Spatial Weights From File spatial conceptualizations are selected. | Double |
Weights_Matrix_File (Optional) | The path to a file containing weights that define spatial, and potentially temporal, relationships among features. | File |
Apply_False_Discovery_Rate__FDR__Correction (Optional) |
| Boolean |
Number_of_Permutations (Optional) | The number of random permutations for the calculation of pseudo p-values. The default number of permutations is 499. If you choose 0 permutations, the standard p-value is calculated.
| Long |
Code sample
ClusterandOutlierAnalysis example 1 (Python window)
The following Python window script demonstrates how to use the ClusterandOutlierAnalysis tool.
import arcpy
arcpy.env.workspace = "c:/data/911calls"
arcpy.ClustersOutliers_stats("911Count.shp", "ICOUNT","911ClusterOutlier.shp",
"GET_SPATIAL_WEIGHTS_FROM_FILE","EUCLIDEAN_DISTANCE",
"NONE","#", "euclidean6Neighs.swm","NO_FDR", 499)
ClusterandOutlierAnalysis example 2 (stand-alone Python script)
The following stand-alone Python script demonstrates how to use the ClusterandOutlierAnalysis tool.
# Analyze the spatial distribution of 911 calls in a metropolitan area
# using the Cluster-Outlier Analysis Tool (Anselin's Local Moran's I)
# Import system modules
import arcpy
# Set property to overwrite outputs if they already exist
arcpy.env.overwriteOutput = True
# Local variables...
workspace = r"C:\Data\911Calls"
try:
# Set the current workspace
# (to avoid having to specify the full path to the feature classes each time)
arcpy.env.workspace = workspace
# Copy the input feature class and integrate the points to snap
# together at 500 feet
# Process: Copy Features and Integrate
cf = arcpy.CopyFeatures_management("911Calls.shp", "911Copied.shp")
integrate = arcpy.Integrate_management("911Copied.shp #", "500 Feet")
# Use Collect Events to count the number of calls at each location
# Process: Collect Events
ce = arcpy.CollectEvents_stats("911Copied.shp", "911Count.shp", "Count", "#")
# Add a unique ID field to the count feature class
# Process: Add Field and Calculate Field
af = arcpy.AddField_management("911Count.shp", "MyID", "LONG", "#", "#", "#", "#",
"NON_NULLABLE", "NON_REQUIRED", "#",
"911Count.shp")
cf = arcpy.CalculateField_management("911Count.shp", "MyID", "!FID!", "PYTHON")
# Create Spatial Weights Matrix for Calculations
# Process: Generate Spatial Weights Matrix...
swm = arcpy.GenerateSpatialWeightsMatrix_stats("911Count.shp", "MYID",
"euclidean6Neighs.swm",
"K_NEAREST_NEIGHBORS",
"#", "#", "#", 6)
# Cluster/Outlier Analysis of 911 Calls
# Process: Local Moran's I
clusters = arcpy.ClustersOutliers_stats("911Count.shp", "ICOUNT",
"911ClusterOutlier.shp",
"GET_SPATIAL_WEIGHTS_FROM_FILE",
"EUCLIDEAN_DISTANCE", "NONE",
"#", "euclidean6Neighs.swm", "NO_FDR", "499")
except arcpy.ExecuteError:
# If an error occurred when running the tool, print out the error message.
print(arcpy.GetMessages())
Environments
Licensing information
- ArcGIS Desktop Basic: Yes
- ArcGIS Desktop Standard: Yes
- ArcGIS Desktop Advanced: Yes
Related topics
- Modeling spatial relationships
- What is a z-score? What is a p-value?
- Spatial weights
- An overview of the Mapping Clusters toolset
- Spatial Autocorrelation (Global Moran's I)
- Hot Spot Analysis (Getis-Ord Gi*)
- How Cluster and Outlier Analysis (Anselin Local Moran's I) works
- Optimized Hot Spot Analysis
- Incremental Spatial Autocorrelation
- Calculate Distance Band from Neighbor Count
- Collect Events
- Optimized Outlier Analysis